Exposome Analytics Course
Terms of Agreement all the code, content, and information is provided "as is". More specifically it is being released under the MIT license. We only ask that you contact us if you decide to use or expand on the work.
I wonder...
How do environmental exposures influence aging?
Spoiler alert Patel CJ, et al. "Systematic correlation of environmental exposure and physiological and self-reported behaviour factors with leukocyte telomere length" Int. J. Epidemiol. (2016) [ PMID: 27059547 | doi: 10.1093/ije/dyw043 | PDF: ]
Download
Description
Slides were produced in a set of two, the first half was an introduction by Chirag and second half was a hands-on demo by Nam.
Tutorial this is an Rmarkdown (.Rmd) file to be used with Rstudio that walks the user through the xwas analysis step-by-step. The html file is a successful run of the tutorial.
- xwas library [ dev | stable ]
- Docker container [ nampho2/xwas ]
- Web Site [ dev | stable ]
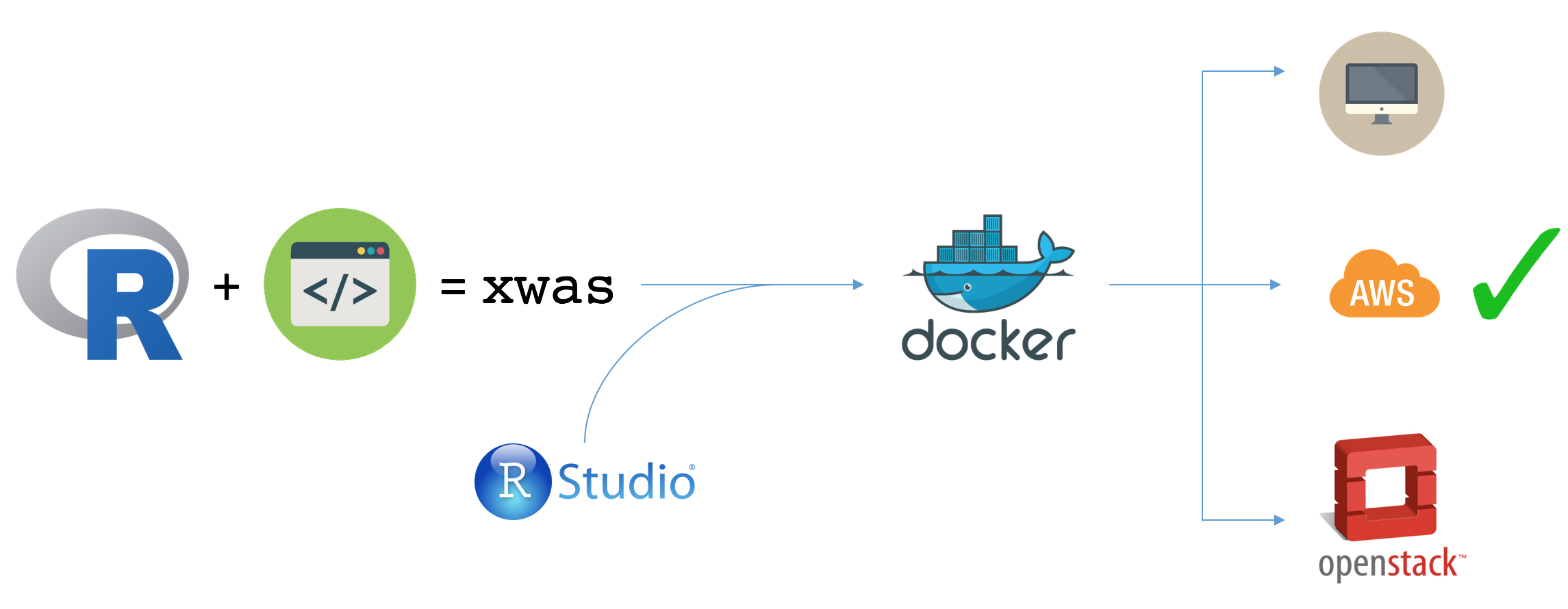
Code the xwas library is under active development as an R package and available to the left. The bleeding edge release of the xwas library with supporting data is packed in a Docker container for this workshop. Source code for this website is also available.
Pre-reqs depending on how you want to experience the tutorial you have some options on which pre-requisites to install.
If you use your own computer you will need to install R at a minimum. If you prefer the more explicit walk through you should install Rstudio (default to this if you aren't sure). If you just install the base R then you can use the demo.html file to guide you through and copy-and-paste the commands. You will need to install the xwas library by following instructions below.
If you want to try the packaged demo then you can use our Docker container locally. You won't have to install the xwas library but you will have to install the Docker tools (link on the left).
If you want to try the packaged demo using Docker but deploy it in the cloud like AWS then you can install the CLI tools (or use the AWS web console) then try online.
Setup your infrastructure
I have R or Rstudio installed and want to work locally
You will have to install the xwas package from Github within R using the devtools library.
library(devtools)
devtools::install_github("nampho2/xwas")
Alternatively you can navigate to the Github source code and there are instructions on how to download the repository and do a compile of the code locally if you are an advanced user. However you get the code installed you can then follow the demo.html (or demo.Rmd if you are using Rstudio) files.
I want to use Docker locally
Follow the links above to download and install Docker on your computer. Once you have it installed you can run the command below from your terminal prompt.
docker run -d -p 80:8787 nampho2/xwas
It instructs Docker to download the nampho2/xwas container that has the libraries and data bundled together. It will then run the service locally on your computer. If you run docker-machine ip you can get the ip address of the virtual machine. If you navigate to that in the browser you'll be able to continue the demo using the Rmarkdown tutorial file.
I want to use AWS
Under construction
Chirag is an assistant professor at Harvard Medical School. Nam is a data scientist in Chirag's lab. Together we are going to show you how to hack the exposome. All content provided here is "as is" and being released under the MIT license. We only ask that you reach out to us before you plan to use or expand on this work.
Citing this work
If you find the data and course materials useful, please cite:
Patel CJ, Pho N, et al. "A database of human exposomes and phenomes from the US National Health and Nutrition Examination Survey" Scientific Data (2016) [ PMID: PMC5079122 | doi: 10.1038/sdata.2016.96 | PDF: ]
Thank You
We would like to acknowledge the organizations below that have supported our work.