Exposure-wide associations to map exposures to phenotypes
Data Science Approaches to Tackle the Complexity of Exposomics: Data, Use Cases, and Models
Introduction
The field of exposomics represents a transformative approach in understanding how environmental exposures influence human health. Traditional paradigms of environmental health, which focus on single exposure-outcome relationships, are evolving to incorporate complex, multifaceted exposures and their interactions with genetic factors. This essay explores data science methodologies and their application to exposomics, emphasizing key questions, models, and the need for comprehensive data resources.
Environmental Health and Exposomics
Environmental health has traditionally centered on identifying causality and risk associated with single exposures. However, the exposome encompasses the totality of environmental exposures from conception onwards, necessitating advanced approaches to understand its impact on health. The exposome includes diverse modalities such as chemicals, diet, lifestyle, and socioeconomic factors, all of which interact in complex ways.
Key Questions in Exposomic Research
- How much variation in disease can be attributed to environmental exposures?
- What specific factors of the exposome are associated with disease?
These questions drive the need for robust data science approaches to parse out the contributions of shared and non-shared environmental factors, gene-environment interactions, and the aggregate effect of multiple exposures.
Data Science Resources for Exposomics
To address these questions, exposomic research requires extensive cohort data that include measures of the exposome, genome, and phenome. Large-scale studies like NHANES and the UK Biobank provide rich datasets for analyzing the complex interactions between genes and the environment.
Exposome-Wide Association Studies (ExWAS)
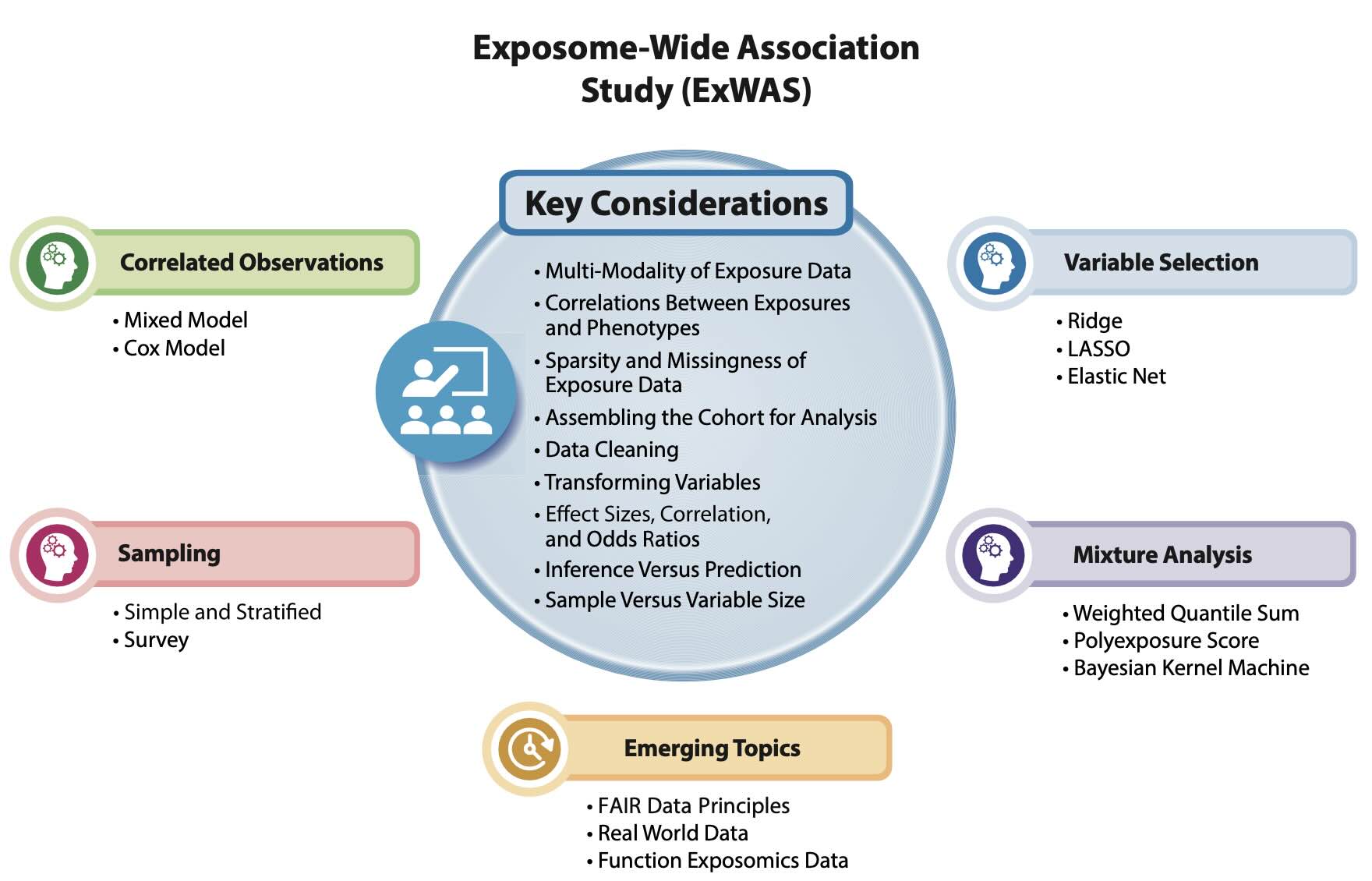
ExWAS is a pivotal methodology in exposomics, analogous to genome-wide association studies (GWAS) in genetics. It systematically examines the relationships between numerous environmental exposures and health outcomes. This data-driven approach allows for the identification of significant associations while controlling for multiple comparisons, making it a powerful tool for discovery in exposomics.
The Complexity of the Exposome
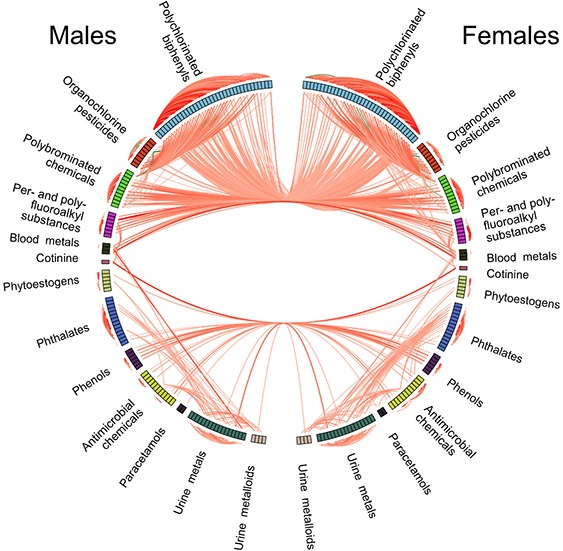
The exposome’s complexity arises from its time-dependent, correlated, and interactive nature. Different modalities of exposure, such as chemicals, physical activity, and socioeconomic status, must be considered collectively to understand their impact on health. The architecture of the exposome includes both shared and non-shared environmental factors, requiring sophisticated statistical models to disentangle these effects.
Methodological Advances
Recent advances in data science have enabled the development of models that can handle the complexity of exposomic data. Techniques such as machine learning, regression models, and mixed models are employed to analyze the dense correlation structures and sparsity of exposure data. Additionally, methods to control for false discovery rates and variable selection are critical to ensuring the robustness of findings.
Exposome-Phenome Atlas
One of the significant advancements in exposomics is the creation of an exposome-phenome atlas. This comprehensive database maps the relationships between various exposures and phenotypes, providing a valuable resource for researchers. By performing ExWAS across multiple cohorts, this atlas facilitates the identification of consistent and replicable associations, enhancing our understanding of how environmental factors contribute to disease.
Conclusions
The integration of data science methodologies into exposomic research represents a paradigm shift in environmental health. By leveraging large-scale cohort data and advanced analytical techniques, researchers can uncover the intricate relationships between environmental exposures and health outcomes. This approach not only enhances our understanding of disease etiology but also informs precision public health interventions aimed at reducing the burden of environmental diseases.
Acknowledgements to collaborators and funding support from NIEHS, NIA, NIDDK, and NIAID.